Description:
Chromatin Biochemistry Using Barcoded Nucleosome Libraries
A Versatile Platform to Screen and Identify Epigenetic Signaling Pathways and Drug Targets
Princeton Docket # 12-2822
Researchers at Princeton University have developed a biochemical strategy to decipher chromatin signaling based on the streamlined preparation of DNA-barcoded nucleosome libraries (‘DNLs’) with distinct combinations of PTMs. These histones are accessible through recombinant (for unmodified histone variants) and protein semi-synthesis technology (for chemically pure, modified histones), the key expertise in the Muir lab. In vitro chromatin immunoprecipitation of these libraries treated with purified effectors (readers, writers, erasers) or nuclear extracts is followed by multiplexed DNA-barcode sequencing (in vitro ChIP-Seq).
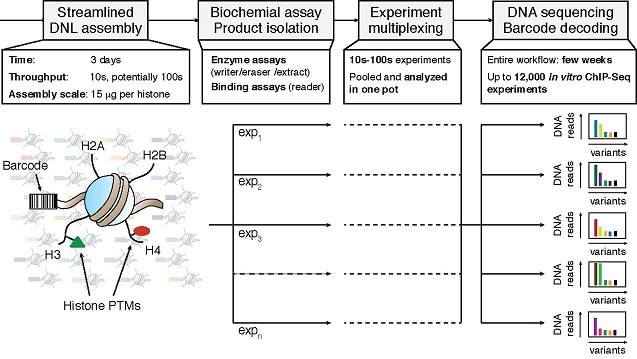
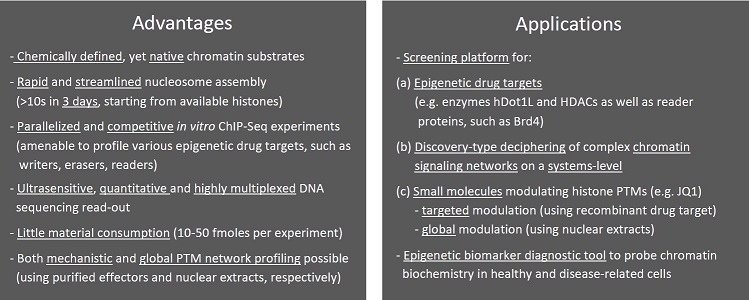
The speed and sensitivity of this workflow enables orders of magnitude higher throughput in nucleosome assembly, biochemical assay read-out and lower material consumption compared to traditional single nucleosome experiments. In their seminal study, the Princeton researchers have prepared a 40-membered nucleosome library to collect many thousands of data points to decipher the mechanisms of how histone PTMs are recognized and deposited by chromatin effectors, including the pharmacologically relevant Bromodomain containing protein 4 (Brd4) and the histone acetyltransferase p300. Importantly, in vitro ChIP-Seq experiments employing the entire writing and reading activities of the nuclear proteome provides a unique opportunity to survey and modulate the complex PTM signal integration network on a systems-level in health and disease
Background
Covalent modifications of histone proteins (posttranslational modifications, ‘PTMs’) and DNA are key processes in the regulation of chromatin. The dynamic installment of PTMs is mediated by ‘writer’ and ‘eraser’ enzymes, while ‘readers’ integrate these (epigenetic) signals into defined transcriptional outputs. The complex interplay of these effectors with chromatin remains poorly understood despite their potential as drug targets for various human pathologies and the body of correlative information generated by top-down ‘omics’ efforts, such as chromatin immune-precipitation (ChIP) and mass spectrometry (MS). Extracting mechanistic details from large-scale datasets requires biochemical approaches that employ chemically defined substrates. Therefore, histone peptide libraries have been extensively used in recent years, but have proven to be poor probes for the identification of inter-histone or histone-DNA dependent regulatory processes. By contrast, nucleosomes (chromatin’s minimal repeating unit) of defined chemical composition provide a more physiological substrate. However, their manufacture and downstream biochemical assays require time- and material-consuming processes. Consequently, nucleosome-based approaches have failed to keep apace with the need for in-depth information on chromatin effectors that is urgently required to develop next generation epigenetic drugs.
Future Directions
The developed approach is now being utilized to identify and characterize a wide array of epigenetic targets, including histone methyltransferases, acetyltransferases and HDACs, as well as to profile the efficacies and specificities of FDA-approved and novel small molecule inhibitors (e.g. JQ1). Additionally, the researchers are further developing the platform for cancer biomarker diagnostics based on misregulation of the identified chromatin signaling networks. While the design and read-out of the current DNL are geared towards investigations of PTM-mediated chromatin processes, the next generations of DNLs (encompassing, e.g., DNA modifications, histone variants and/or mutants) will open up new approaches to understand and target chromatin signal integration networks on a large scale.
Publication: Nguyen, U. T. T., Bittova, L., Müller, M. M., Fierz, B., David, Y., Houck-Loomis, B., Feng, V., Dann, G. P., and Muir, T. W. “Accelerated Chromatin Biochemistry using DNA-Barcoded Nucleosome Libraries.” Nature Methods, NMethod.3022, on line. 6 July 2014.
Inventors: Tom W. Muir, Uyen T.T. Nguyen, Manuel M. Müller
Intellectual property and technology status: Patent pending
The office of technology licensing to looking to identify appropriate partners for the further development and commercialization of this technology
Contacts:
Laurie Tzodikov
Princeton University Office of Technology Licensing • (609) 258-7256• tzodikov@princeton.edu
Tom W. Muir, Professor of Chemistry
Professor Muir received his Ph.D. in organic chemistry in 1993 from the University of Edinburgh. After studying bioorganic chemistry at The Scripps Research Institute, he joined Rockefeller in 1996 as assistant professor, becoming Richard E. Salomon Family Professor in 2005. In 2009, he moved his lab to the Department of Chemistry in Princeton to become the Van Zandt Williams Jr. Class of ’65 Professor of Chemistry. Professor Muir has received the Blavatnik Award for Young Scientists, the Vincent du Vigneaud Award, the Irving Sigal Young Investigator Award, the Leonidas-Zervas Award, and a Burroughs Wellcome Fund New Investigator Award. He was deemed an Alfred P. Sloan Research Fellow in 1999 and a Pew Scholar in the Biomedical Sciences in 1997. Professor Muir is a fellow of the American Association for the Advancement of Science.
Uyen T. T. Nguyen, Post-Doctoral Researcher
Dr Nguyen studied chemistry at the University of Cologne, Germany, where she received her diploma degree with summa cum laude in 2005. In 2009, she obtained her Ph.D. under the supervision of Professor Roger S. Goody and Professor Herbert Waldmann at the Max-Planck-Institute of Molecular Physiology, Dortmund, Germany. During her dissertation, she developed a novel technology to tag, isolate and analyze the entire eukaryotic prenylome. Since then, she has been carrying out postdoctoral research on chromatin biochemistry at Rockefeller University and later Princeton University with Professor Tom W. Muir.
Manuel M. Müller, Post-Doctoral Researcher
Dr Müller obtained a masters degree with distinction in Biochemistry from the ETH Zürich in 2006. After a summer research stay in the laboratory of Professor Andy Ellington at the University of Texas, Austin, he returned to the Laboratory of Organic Chemistry at ETH Zürich to pursue his doctoral studies under the supervision of Professor Donald Hilvert. His PhD thesis involving model systems for primordial enzymes was awarded with the ETH medal in 2011. In 2010, Manuel joined the laboratory of Professor Tom W. Muir, now at Princeton University, where he is developing novel tools for chromatin biochemistry.